Pathology has long been the cornerstone of disease diagnosis, relying on detailed examination of tissue samples to uncover cellular abnormalities and guide treatment decisions. With the rise of digital pathology, Whole Slide Images (WSIs) have emerged as transformative tools, offering ultra-high-resolution, gigapixel-sized digital scans of histopathological tissue slides. These images capture intricate cellular and structural details necessary disease diagnosis, cancer grading, and biomarker identification.
Yet, the sheer size and complexity of WSIs challenge traditional machine learning methods. Models designed for smaller and more uniform inputs (like standard image datasets) struggle to handle the scale and variability of WSIs. Processing these images effectively requires specialized techniques that can extract meaningful, feature-rich representations—without compromising the spatial or biological context critical for clinical insights.
To address the inherent complexity of Whole Slide Images (WSIs), EmPathy, our AI-powered accelerator, leverages a modular deep learning pipeline optimized for scalability, interpretability, and performance in computational pathology. The system is specifically designed to process high-resolution WSIs by breaking them down into manageable, model-compatible components without losing critical contextual information.
- Tiling for Scalability: EmPathy segments gigapixel WSIs into non-overlapping tiles, enabling efficient downstream processing within memory and compute limits.
- Self-Supervised Embedding: Tiles pass through a foundation model trained via self-supervised learning, extracting rich histopathological feature embeddings.
- Tile-Level Classification: A lightweight classifier distinguishes cancerous from non-cancerous tiles with optimized precision and recall.
- Embedding Aggregation & Clustering: Similar embeddings cluster spatially, with weighted averaging generating slide-level representations for fast, semantic search.
- Interactive Visualization: AI-driven overlays highlight cancerous regions on WSIs, enhancing pathologist interpretability and diagnostic efficiency.
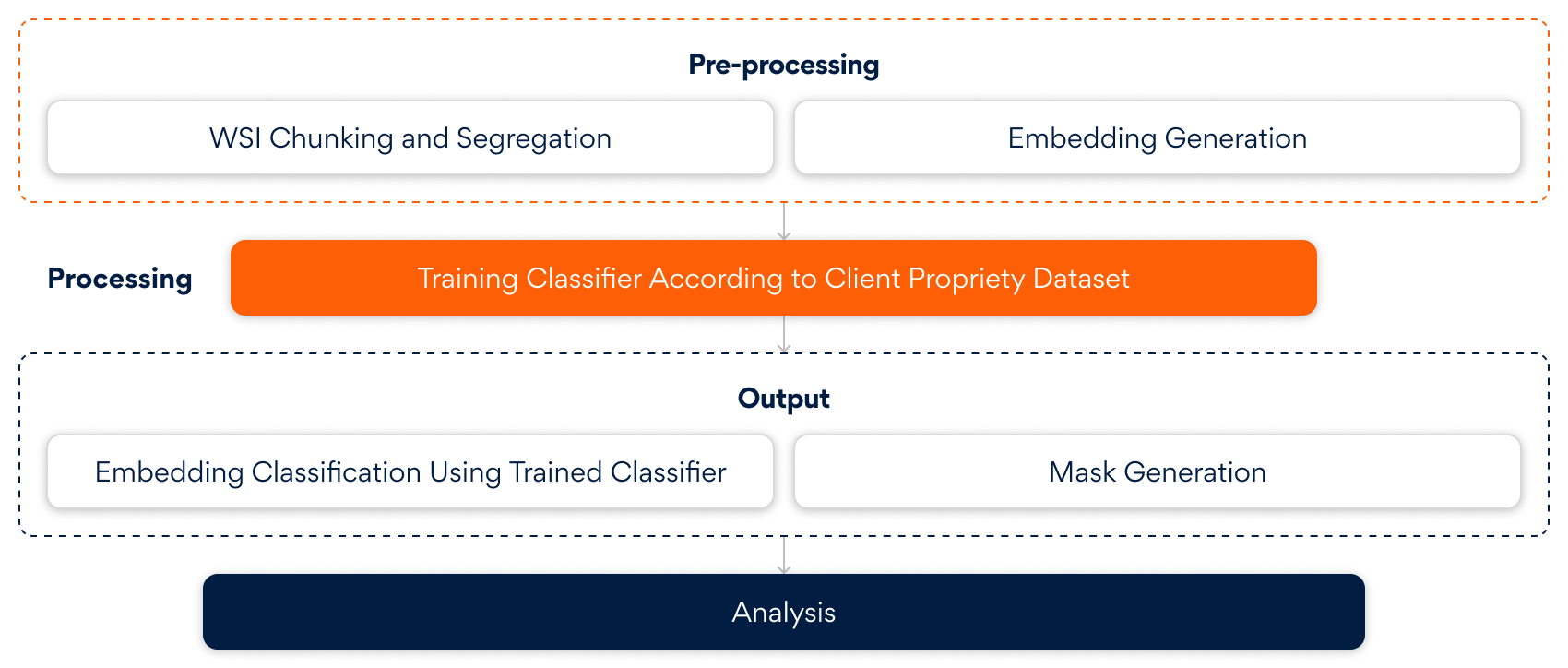
The above diagram illustrates the end-to-end AI-driven pipeline that transforms raw Whole Slide Images (WSIs) into clinically actionable insights. It begins by intelligently breaking down and embedding complex pathology data. Using client-specific datasets, a tailored classifier is trained to detect subtle patterns. The result is a precise classification of embeddings and the generation of spatial masks enabling downstream analysis with contextual clarity and clinical relevance.
Core Intelligence: Foundation Model Embeddings
Empathy leverages a transformer-based vision foundation model, pre-trained on large-scale histopathological datasets, to generate high-dimensional, feature-rich embeddings for each tile. These embeddings capture fine-grained visual patterns such as cellular morphology, tissue architecture, and staining variations—complex cues that are nearly impossible to manually engineer.
Tile Classification: Modular and Efficient
A lightweight classifier, fine-tuned on pathologist-annotated data, operates on fixed foundation model embeddings. This modular setup optimizes both accuracy and efficiency, reducing compute load and enabling scalable, high-throughput deployment across varied pathology tasks.
Clustering for Spatial Insight
Density-based clustering groups similar embeddings across WSIs, ensuring robustness to noise. Aggregated cluster features create compact region-level representations, enhancing interpretability, enabling localized decisions, and lowering computational costs for downstream analysis.
Diagnostic Overlay: Interpretable Cancer Mask
The visualization layer projects prediction scores onto the original WSI, highlighting cancerous clusters with precision shading. This spatial mask enables rapid identification of high-risk regions. Designed for clinical interpretability, it bridges AI-driven inference with pathologist workflows, facilitating faster, more informed diagnostic decisions without sacrificing transparency or spatial context.
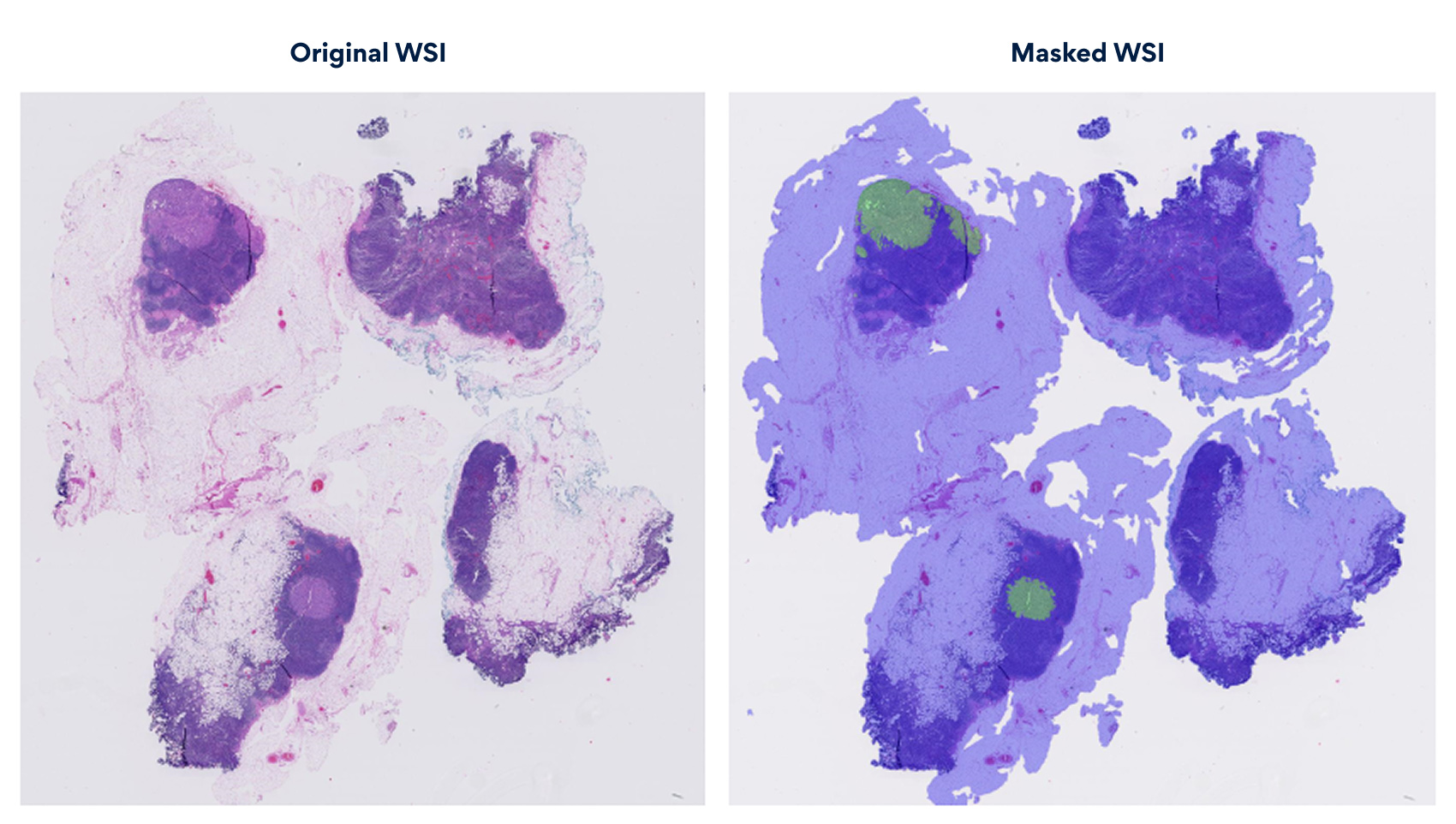
The diagram illustrates the masked WSI, highlighting regions of concern. The masked overlay projects cancerous clusters onto the raw scan, providing a clear, interpretable map. This visualization embeds explainable AI insights at the pixel level, enhancing pathologist decision-making with clarity and transparency.
Business Impact and Value Proposition
Organizations and clinics leveraging AI-powered WSI analysis can unlock significant operational efficiencies, improve diagnostic accuracy, and enhance patient care through scalable, interpretable, and cost-effective solutions.
- Accelerated Diagnostics: Enables rapid, high-throughput analysis, reducing turnaround times by up to 40% and increasing pathology lab efficiency by 30%.
- Enhanced Accuracy: Improves detection of subtle cancerous patterns, minimizing diagnostic errors by approximately 25% and supporting better patient outcomes.
- Scalable Integration: The modular design allows seamless deployment across diverse clinical workflows without heavy computational demands.
- Improved Interpretability: Visual overlays enhance pathologist trust and streamline decision-making with explainable AI insights.
- Cost Efficiency: Reduces manual annotation and review burden, lowering operational costs while maintaining clinical rigor.
EmPathy embodies the fusion of advanced AI and human expertise, delivering not just accuracy but also transparency, reliability, and deep interpretability in WSI analysis. With clear visualization, it empowers pathologists to make confident, informed decisions—transforming gigapixel complexity into actionable insights with purpose and precision.
At Persistent, we are combining human expertise, advanced AI, and biology to enable precise, efficient, and explainable diagnostic insights.
Contact us to get started with Precision Medicine.
Author’s Profile
Shilpa Ramteke
Senior Data Scientist, Innovation Labs AI Research
Akansha Wasalu
Senior Software Engineer, Innovation Labs AI Research








