Traditional Drug Discovery Challenges
Drug discovery and development requires significant financial investment and several years to bring a single drug to the market. Advancements in biomedical sciences are pushing regulatory agencies (such as FDA) to prioritize approval of safer, more selective drugs with fewer side effects and risks for patients.
Traditionally, the practice of drug discovery involves screening several thousands of chemical compounds in an experimental setup to identify a few hit compounds. Over the last two decades, in silico virtual screening has dramatically accelerated the initial search for potential drugs, reducing the need for lab testing to merely a few hundreds. However, the subsequent crucial step – developing one of these promising ‘hits’ into an optimized drug ready for human trials still requires a time-consuming process of physically creating and testing many analogs of the hit molecule.
Deep Learning and Generative AI in Drug Discovery
The development of the revolutionary and Nobel prize-winning AlphaFold2 model for protein structure prediction has marked a significant breakthrough in AI applications within the field of life sciences. Since then, large predictive and generative AI models specialized for biology and chemistry-related tasks have been revolutionizing the drug discovery and biomedical R&D landscape. These state-of-the-art AI models help scientists to execute tasks like protein structure predictions, novel molecule design, molecular property predictions and modeling molecular interactions with superior speed, accuracy and broader applicability compared to physics-based methods.
Drug discovery across pharma and biotech industries face challenges for AI adoption, integration and deployment, primarily due to the complex nature of these models requiring specialized skills, fast evolving nature of the technology, high computation resources and need for fine tuning of these models on specific data.
Re(AI)magining Drug Discovery with GenMolVS
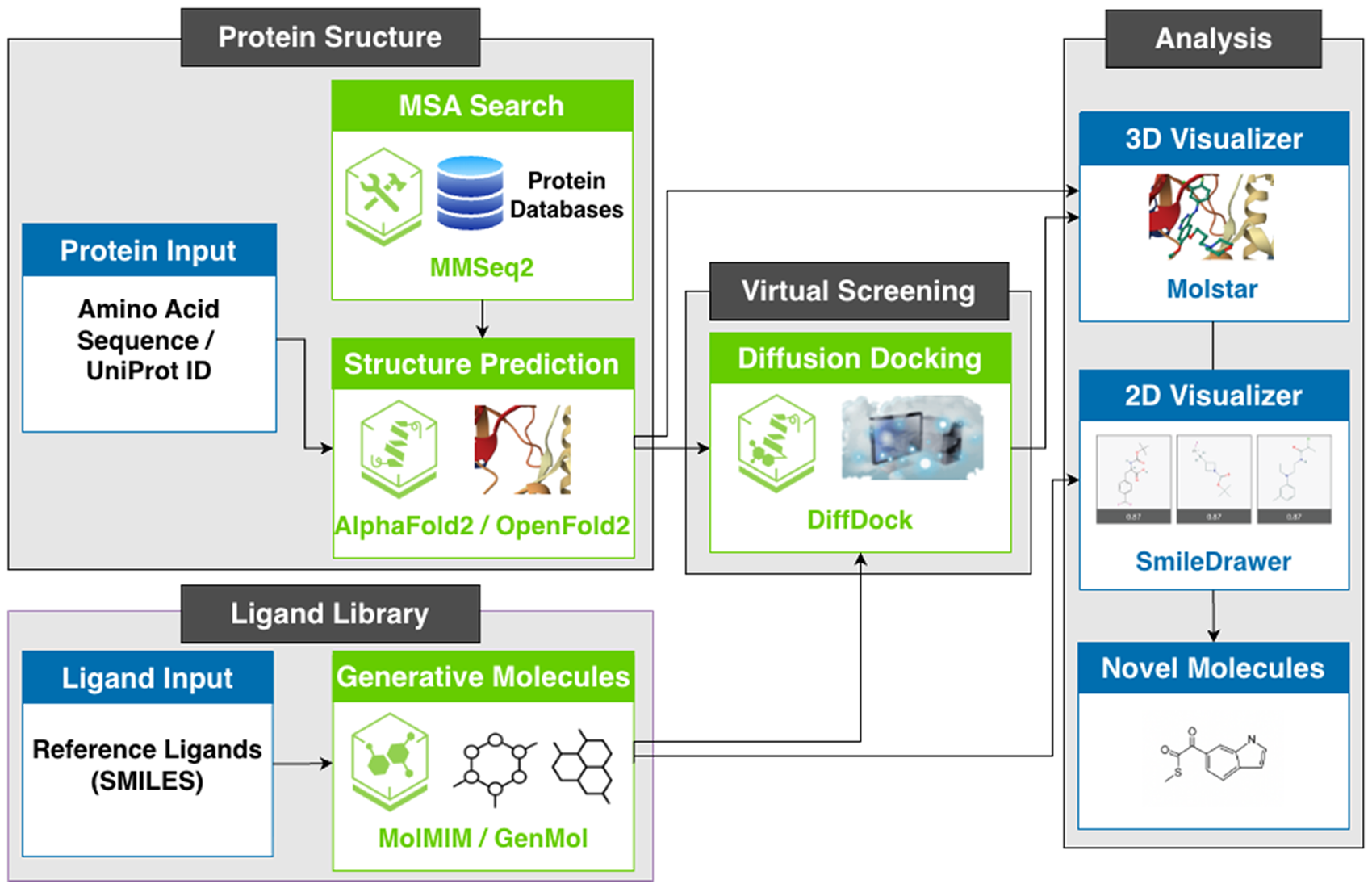
Generative Molecules & Virtual Screening (GenMolVS) pipeline is an integrated solution developed by Persistent Systems by leveraging NVIDIA BioNeMo NIMs that encapsulate pretrained and fine-tuned deep learning and generative AI models. The pipeline enables scientists to accelerate several major critical tasks of preclinical drug discovery.
GenMolVS provides a seamless digital workflow that starts with the sequence of a disease-associated gene or protein and leads to the selection of a very few potent drug-like molecules. Specifically, GenMolVS redefines three major tasks of the preclinical R&D:
- Accurate prediction of a protein’s 3D structure, thereby reducing reliance on experimental methods.
- De Novo design of novel molecules with optimized properties, thereby minimizing the need for high-throughput screening of extensive chemical libraries and large number of molecule synthesis.
- Generative docking to simulate ligand-protein interactions, allowing prioritization of fewer but high-confidence molecules that are worth testing experimentally.
This GenAI solution is anticipated to significantly shorten the preclinical drug discovery process, enabling clinical trials to commence within two years instead of the typical 5-to 6-year timeframe.
Target Protein Structure Prediction
Having accurate 3-dimensional (3D) structures of therapeutic target proteins is the first and foremost task in drug discovery. Traditionally, scientists across academics and industry must spend years synthesizing and purifying proteins to obtain experimental structures of proteins. The first module, ‘Protein Structure Prediction’ of GenMolVS allows scientists to predict 3D structures of therapeutically relevant proteins using a set of leading deep learning models like AlphaFold2 or OpenFold2.
Depending on the use-case or specific requirements, scientists can use the most suitable model for protein structure predictions. For instance:
AlphaFold2: for predicting protein structures with near experimental accuracy.
OpenFold2: for innovation requiring more accessible and customizable models. In addition, OpenFold2 can use proprietary protein structures of similar sequences as templates for improved prediction accuracy.
Thus, providing choices of multiple models gives the freedom to accurately and efficiently predict the protein structure that suits different use cases across industry.
Beyond Searching – Designing Novel Molecules with Generative AI
The second module ‘Molecule Generation’ empowers scientists with two generative models – Mutual Information Machine (MIM) learning (MolMIM) and GenMol developed by NVIDIA for designing novel molecules with optimized and desired qualitative estimates of drug-likeness (QED) properties which are crucial for success in later stage of clinical trials.
- MolMIM: leverages a probabilistic auto-encoder to generate novel molecules with desired properties starting with known hits or lead compounds as reference molecules.
- GenMol: employs a discrete diffusion model to generate molecules piece-by-piece using molecular fragments. This fragment-based approach makes it versatile for various complex drug designs and lead optimization tasks such as scaffold morphing / decoration, and fragment remasking, etc.
Altogether, MolMIM and GenMol address the need for generating novel molecules and optimizing lead molecules, which would likely succeed in the subsequent clinical phases.
Beyond Traditional Docking: Achieving Higher Accuracy and Speed with Generative Docking
The third module of the pipeline uses a diffusion generative model DiffDock for generative molecular docking. Molecular docking is important for determining the likelihood of a chemical ligand interacting with a disease protein and inferring its drug effect.
Traditional docking methods commonly used by the drug discovery industry require higher computation times but achieve only low to moderate accuracy. DiffDock is a contemporary model for generating binding poses between small molecules and proteins with improved accuracy and speed compared to other generative as well as physics-based docking methods. This enables a better understanding of potential molecular interactions with high confidence estimates and accuracy.
At the end of the pipeline, scientists can select a few highly potent molecules that can be quickly synthesized and screened in laboratory studies. GenMolVS will be extended with more capabilities and features for maximizing the chance of success of molecules in clinical trials. Together with NVIDIA, the solution can be rapidly adapted to the ever-emerging and evolving needs specific to the pharma industry.
Business Impact
- Time and Cost Reduction: By reducing the reliance on expensive and time-consuming processes of experimental structures and by designing potent drug-like molecules, GenMolVS pipeline can potentially shorten the timeline and reduce the cost for drug development.
- Higher success rate of drug discovery: Higher accuracy of each of the module of GenMolVS pipeline would mean higher success rate at clinical trial allowing companies to bring new drugs to market faster, potentially increasing revenue and profitability.
- Competitive Advantage: Leveraging state-of-the-art AI models integrated into scalable and flexible pipelines provides a competitive edge in the pharmaceutical and biotech industries. Companies that adopt these solutions can strengthen their position, expand their therapeutics pipelines and stay ahead of their competitors by developing safer, more selective medicines with fewer side effects.
Accelerate Drug Discovery with NVIDIA BioNeMo Microservices
NVIDIA BioNeMo NIM are specialized microservices designed to accelerate AI-driven drug discovery. They are provided as ready-to-deploy containers, each bundling a specific AI model with all its dependencies. Featuring pre-optimized inference engines, these NIM leverage the massive parallel processing power of NVIDIA GPUs to drastically reduce the time needed for complex computational simulations.
These microservices can be deployed in various environments like cloud, data centers and even personal workstations. They provide industry-standard APIs that can be seamlessly plugged into any customizable and scalable workflow, reducing the complexity of setting up the advanced AI models. NVIDIA NIM also ensure compliance with any data governance requirements.
Persistent Systems brings deep domain knowledge, AI/ML proficiency and engineering expertise to build specialized AI applications for the biopharma and biotech industries. The unique project requirements may extend beyond standard NIM requiring custom features and capabilities. Based on the need for tailored solutions, Persistent can utilize the NVIDIA BioNeMo Framework to fine-tune AI models using client proprietary data. Our close collaboration with NVIDIA ensures we deliver adaptable, cutting-edge AI solutions that rapidly meet the evolving needs of pharmaceutical companies.
Conclusion
Persistent’s GenAI solution, GenMolVS, enables scientists to accurately and efficiently predict protein’s 3D structure, screen chemical libraries or design novel drug molecules with optimized properties and predict their binding to the target protein. This process facilitates the selection of the most promising molecules with high confidence for synthesis and experimental validation. Built with pre-optimized AI models and high-performance computing from NVIDIA, GenMolVS empower pharma companies to accelerate the discovery of life-saving medicines, for almost all protein targets thereby designing the keys to unlock future therapies more efficiently than ever before.
GenMolVS will soon feature agentic AI capabilities—that is, AI systems endowed with reasoning and decision-making ability. These agents will execute tasks such as analyzing a researcher’s request for design and selection of optimal drug candidates against a target by autonomously running simulations across various AI models until they discover the most promising molecule for synthesis and validation. This approach would ensure that domain-specific AI models are utilized in a highly optimized manner, ultimately leading to faster and more robust solutions for real-world drug discovery challenges.
Author’s Profile
Dr. Som Dutt
Principal Domain Expert, HLS, Persistent Systems
Leena Bahulekar
Senior Technical Consultant, Persistent Systems








